Introduction
By introducing the File object in EPrints 3.2 we are able to directly manipulate the physical objects (files) which are related to our ePrints. In this way analysis of particular files can be performed in order to analyse any risks affecting our repository.
Secondly, having a breakdown of the file types in your repository has also been proved to be useful through the popularity of services such as roar
For further background information on this topic:
- Presentation - EPrints and Preservation, In: Tackling the Preservation Challenge: Practical Steps for Repository Managers, 12th December 2008, London
- Publication - Where the Semantic Web and Web 2.0 meet format risk management: P2 registry, In: iPres2009: The Sixth International Conference on Preservation of Digital Objects
Tutorial - Aim
Aim: The aim of this tutorial is to introduce some of the new features of EPrints 3.2 by using DROID (which is hidden under EPrints) to classify some objects in our repository. Further to this we then look at "risk scores" associated with these formats which leads us to introduce this part of current research in digital preservation.
Note: At time of writing (September 2009), the National Archives (UK) PRONOM service was not yet providing risk scores and thus a demonstration service is used in part of this tutorial.
This tutorial is written specifically for a set of demonstration repositories and some parts of the interface are not part of the standard EPrints 3.2 release.
File Classification
In this section we look at classifying the files in your repository using DROID from droid.sourceforge.net/ and the classification add-ons available from files.eprints.org. In the tutorial repositories both of these packages have already been installed, to install them on your repository download the classification add-ons from files.eprints.org and follow the README instructions on how to install them as well as DROID.
To classify files in a live repository it is recommended that the process is run using a scheduled job which runs at most a couple of times a day, for the purposes of the tutorial a button has been provided in the admin interface which invokes it on demand.
The rest of this tutorial is split into exercises applicable to the tutorial and those applicable in all cases.
The Formats/Risks Screen
This screen will be our main reference point throughout the exercise. Available via the admin interface (show below) we can view the file types in our repository and any related risk scores.
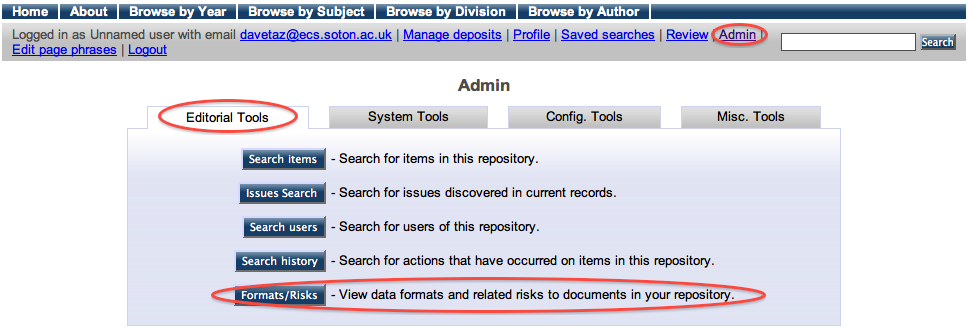
Viewing this page from an empty repository should result in the following screen.
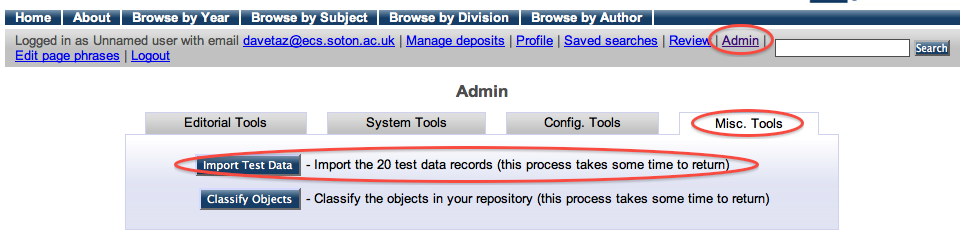
Exercise 1 - Populating the repository (tutorial repositories only)
For the purposes of this tutorial, we have provided a set of 20 records from the EPrints test dataset. To import these an Import Test Data button is available from the Misc. Tools section of the Admin interface. This process takes some time to return, please be patient.
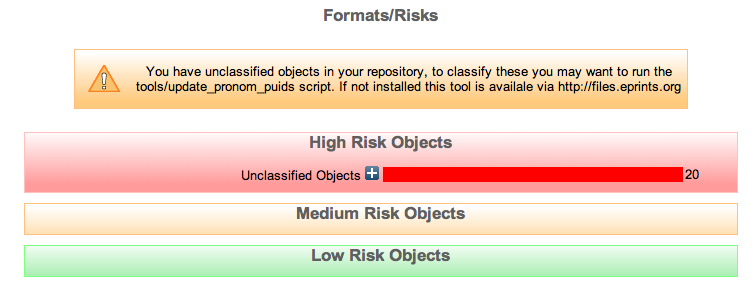
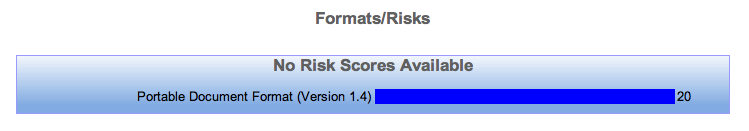
After this process is finished if we browse back to the Format/Risks Screen we should see that there are 20 unclassified objects.
Exercise 2 - Classifying the objects in your repository
Non-TutorialFrom the command line, as the eprints user, the following command needs to be run in order to classify the objects in your repository:
The classification process can be performed through the Classify Objects button available via the Misc. Tools tab on the Admin interface.
This button performs exactly the same command as that called by the non-tutorial users, except it requires no command line access. The downside of this is that it is not run in the background on a regular basis.
As a result of the above process our Format/Risks Screen should now be showing classified objects. The below screenshot is applicable only to the tutorial.
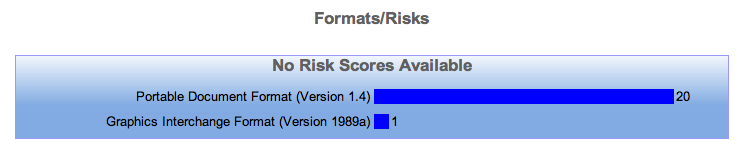
Exercise 3 - Adding an "at risk" example file (Tutorial Only)
As part of the tutorial we have provided a set of files available here, uploading one of these to the repository and then repeating the classification process at the end of exercise 2 should lead us to to a repository profile much like the one shown below.
Risk Analysis (Exercise 4)
To enable risk analysis we need edit the config file for PRONOM available via the View Configuration button available from the Config. Tools tab of the admin interface. the pronom.pl config file is near the top in the first cfg.d section.
A more detailed guide on this step can be found in the Code Changing guide.
Note: Please be careful when changing this file, this feature is not enabled normally via the web interface and requires command line access.In this file we want to edit the $c->{"pronom_unstable"} = 0; and replace the 0 with a 1 to read $c->{"pronom_unstable"} = 1;.
Finally we need to follow the Code Changing guide to reload the configuration and then Classify the objects to update to the new risk scores which are now supplied from a different service.
This should now lead to the Format/Risks Screen displaying the following, showing that the previously added gif format is high risk.
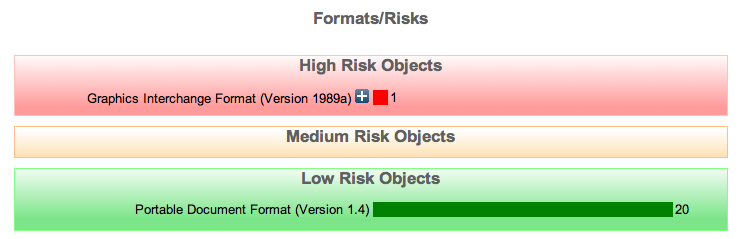
The Planets Project part of this tutorial will go further to explain what can be done with potential high risk formats. Also the intro presentation should have gone some way to help explain the importance of file formats in digital preservation.
Exercise 5 - Moving Risk Boundaries and Changing Re-Classification Period
The configuration file we edited in the last section can also be used to control the risk score boundaries between high, medium and low risk. The National Archives (UK) schema is to provide a score based on 8 classification categories which is between 0 and 3000. Thus for simplicity the default boundaries have been set at 0-1000 for high risk, 1000-2000 for medium risk and 2000+ for low risk. By moving these to be 0-100,100-200 and 200+ respectively you should be able to change the classification of the gif risk score. In the configuration file these parameters are listed by default as $c->{"high_risk_boundary"} = 1000; and $c->{"medium_risk_boundary"} = 2000;
Finally this configuration file also allows you to change how often a re-classification of files is performed. Defined in seconds the default value is 30 days as detailed in the configuration file by the line: $c->{pronom}->{max_age} = 30 * 86400; # 30 days